Kernel-MMD-Hypothesis Testing without assumptions
What to do if you can't assume anything about your distributions.
Introduction
The basic problem in statistics is comparing two samples of measurements. The basic issue is that you want to know whether these measurements came from the same source, or from different sources. In statistical language, we are comparing the null distribution—that the measurements came from the same source—against the alternative hypothesis —that the measurements come from distinct sources.
Very often statisticians assume some parametric form for the distributions. For example, normal distributions with known unknown means and the same variance. So we would be comparing:
\[H_0: \mu_1 = \mu_2\]Against
\[H_0: \mu_1 \neq \mu_2\]There are some downsides to this approach. Firstly, we are imposing an assumption on the data—that the measurements come from a distribution with a specific form—and we can never check the accuracy of that assumption. That is if we have gotten the correct parametric form the distributions. Secondly, the specific distributional form we assume is also unknowable. So we can never really know how wrong we are.
The issue then is to find solutions to the following problem, without making assumptions about the data generating process of the measurements.
Problem 1 Let $x$ and $y$ be random variables defined on some topological space $\mathcal{X}$, with respective probability measures $p$ and $q$. Given two sets of observations $X = \left(x_1, x_2, \ldots, x_m \right)$ and $Y = \left(_1, y_2, \ldots, y_n \right)$ drawn i.i.d from $p$ and $q$ respectively, can we decide whether $p \neq q$?
Maximum Mean Discrepancy (MMD)
In general, MMD is defined by the idea of representing distances between distributions as distances between mean embeddings of features. We can define the notion of distance between probability distirbutions with the following lemma (from (Dudley, 2018)).
Lemma 1 Let $\left(\mathcal{X}, d\right)$ be a metric space, and let $p$ and $q$ be two probability measures defined on $\mathcal{X}$. Then $p = q$ if and only if $\mathcal{E}_x\left(f\left(x\right)\right) = \mathcal{E}_y\left(f\left(y\right)\right)$ for all $f \in C\left(\mathcal{X}\right)$ where $C\left(\mathcal{X}\right)$ is the collection of bounded, continuous functions on $\mathcal{X}$.
The maximum mean discrepancy (MMD) is then defined based on a this class of functions:
Definition 1 Let $\mathcal{F}$ be a class of functions $f: \mathcal{F} \rightarrow \mathcal{R}, and let $p, q, x, y, X, Y$ be defined as earlier. The MMD is then:
\[MMD(p,q) := \mathrm{sup}_{f \in \mathcal{F}} \left( \mathcal{E}_x[f(x)] - \mathcal{E}_y[f(y)] \right)\]An empirical estimate of the MMD can be found by replacing the population expectations with their sample expectations based on the samples $X$ and $Y$ (although this is slightly biased):
\[\hat{MMD(p,q)} := \mathrm{sup}_{f \in \mathcal{F}} \left( \frac{1}{m}\sum_{i=1}^m f(x_i) - \frac{1}{n}\sum_{i=1}^n f(y_i) \right)\]Although $C\left(\mathcal{X}\right)$ allows us to identify if $p = q$, the space is too rich. It is not computationally practical to work in this space. Instead we will work with a function class which can identify whether $p = q$ but is restrictive enough to provide useful estimates with finite samples.
Reproducing Kernel Hilbert Spaces
A Reproducing Kernel Hilbert Space (RKHS) is a space $\mathcal{X}$ of functions, equipped with a norm (i.e. a Hilbert space). There is a function $\phi(x)$ which takes points in $\mathcal{X}$ to $\mathcal{R}$. We denote this function $ f(x) = \langle f, \phi(x) \rangle$. We can write $\phi(x) = k(x, \dot)$. In particular
\[k(x, y) = \langle \phi(x), \phi(y) \rangle\]Speaking informally, an RKHS is just a space where every point in the space is a linear combination of (positive-definite) kernels. This allows us to replace the inner product calculation in this space with the kernel evaluation. We can, in particular, extend the notion of a feature map to the embedding of a probabuility distribution. Let $ \mu_p \in \mathcal{H}$ be such that $\mathcal{E}x[f] = \langle f, \mu_p \rangle$. We call this the _mean embedding of $p$. Then, the MMD may be expressed as the distance between mean embeddings in $\mathcal{H}$/
Kernel MMD
$\phi:\mathcal{X} \rightarrow \mathcal{H}$, where $\mathcal{H}$ is some Hilbert space; this corresponds to a kernel by $k\left(x,y\right)=\left⟨\phi(x),\phi(y)\right⟩$. The MMD is:
\[MMD^2\left[p, q \right] = \left\lVert \mu_p - \mu_q \right\rVert^2\]And we can obtain the MMD in terms of the RKHS kernel functions:
\[MMD^2\left[p, q \right] = \mathcal{E}_{x, x'}[k(x, x')] - 2\mathcal{E}_{x, y}[k(x, y)] + \mathcal{E}_{y, y'}[k(y, y')]\]This is relatively easy to compute (implmentation from torchdrift):
import torch
def mmd(x, y, sigma):
# compare kernel MMD paper and code:
# A. Gretton et al.: A kernel two-sample test, JMLR 13 (2012)
# http://www.gatsby.ucl.ac.uk/~gretton/mmd/mmd.htm
# x shape [n, d] y shape [m, d]
# n_perm number of bootstrap permutations to get p-value, pass none to not get p-value
n, d = x.shape
m, d2 = y.shape
assert d == d2
xy = torch.cat([x.detach(), y.detach()], dim=0)
dists = torch.cdist(xy, xy, p=2.0)
# we are a bit sloppy here as we just keep the diagonal and everything twice
# note that sigma should be squared in the RBF to match the Gretton et al heuristic
k = torch.exp((-1/(2*sigma**2)) * dists**2) + torch.eye(n+m)*1e-5
k_x = k[:n, :n]
k_y = k[n:, n:]
k_xy = k[:n, n:]
# The diagonals are always 1 (up to numerical error, this is (3) in Gretton et al.)
# note that their code uses the biased (and differently scaled mmd)
mmd = k_x.sum() / (n * (n - 1)) + k_y.sum() / (m * (m - 1)) - 2 * k_xy.sum() / (n * m)
return mmd
(Gretton et al., 2012) recommends to set the $\sigma$ parameter to the median distance between points:
\[\sigma = \frac{\mathrm{Median}\left(z_i - z_j\right)}{2}\]where $Z$ is the combined sample of and $X$ and $Y$. We have also used the Gaussian Radial Basis as the choice of kernel.
We can extend this implementation to any kernel with the following code:
import numpy as np
from scipy.spatial.distance import pdist, squareform
from scipy import linalg
from typing import Union, Tuple, List
def mmd_biased(XX: np.ndarray, YY: np.ndarray, XY: np.ndarray) -> float:
"""
Compute biased MMD^2 statistic.
Args:
XX: Kernel matrix for first sample
YY: Kernel matrix for second sample
XY: Cross kernel matrix between samples
Returns:
float: Biased MMD^2 statistic
"""
m = XX.shape[0]
n = YY.shape[0]
return (np.sum(XX) / (m**2)) + (np.sum(YY) / (n**2)) - (2 / (m*n)) * np.sum(XY)
def mmd_unbiased(XX: np.ndarray, YY: np.ndarray, XY: np.ndarray) -> float:
"""
Compute unbiased MMD^2 statistic.
Args:
XX: Kernel matrix for first sample
YY: Kernel matrix for second sample
XY: Cross kernel matrix between samples
Returns:
float: Unbiased MMD^2 statistic
"""
m = XX.shape[0]
n = YY.shape[0]
term1 = (np.sum(XX) - np.trace(XX)) / (m * (m-1))
term2 = (np.sum(YY) - np.trace(YY)) / (n * (n-1))
term3 = (2 / (m*n)) * np.sum(XY)
return term1 + term2 - term3
def mmd2test(K: np.ndarray, label: Union[List, np.ndarray],
method: str = "b", mc_iter: int = 999) -> dict:
"""
Kernel Two-sample Test with Maximum Mean Discrepancy.
Maximum Mean Discrepancy (MMD) as a measure of discrepancy between samples
is employed as a test statistic for two-sample hypothesis test of equal
distributions. Kernel matrix K is a symmetric square matrix that is positive
semidefinite.
Args:
K: Kernel matrix (symmetric, positive semidefinite)
label: Label vector of class indices
method: Type of estimator to be used. "b" for biased and "u" for unbiased
mc_iter: Number of Monte Carlo resampling iterations
Returns:
dict: Dictionary containing test results with keys:
- statistic: Test statistic
- p_value: p-value under H0
- alternative: Alternative hypothesis
- method: Name of the test
"""
# Preprocessing
K = np.asarray(K)
if not (K.ndim == 2 and K.shape[0] == K.shape[1]):
raise ValueError("K should be a square matrix")
if not np.allclose(K, K.T):
raise ValueError("K should be symmetric")
# Check if K is positive semidefinite
min_eigenval = np.min(linalg.eigvalsh(K))
if min_eigenval < 0:
print(f"Warning: K may not be PD. Minimum eigenvalue is {min_eigenval}")
# Process labels
label = np.asarray(label)
unique_labels = np.unique(label)
if len(unique_labels) != 2:
raise ValueError("label should contain exactly 2 classes")
if len(label) != K.shape[0]:
raise ValueError("Length of label must match size of kernel matrix")
# Compute statistic
id1 = np.where(label == unique_labels[0])[0]
id2 = np.where(label == unique_labels[1])[0]
m, n = len(id1), len(id2)
if method.lower() == "b":
stat = mmd_biased(K[np.ix_(id1, id1)], K[np.ix_(id2, id2)], K[np.ix_(id1, id2)])
else: # method == "u"
stat = mmd_unbiased(K[np.ix_(id1, id1)], K[np.ix_(id2, id2)], K[np.ix_(id1, id2)])
# Monte Carlo iterations
iter_vals = np.zeros(mc_iter)
for i in range(mc_iter):
perm = np.random.permutation(m + n)
tmp_id1 = perm[:m]
tmp_id2 = perm[m:]
if method.lower() == "b":
iter_vals[i] = mmd_biased(K[np.ix_(tmp_id1, tmp_id1)],
K[np.ix_(tmp_id2, tmp_id2)],
K[np.ix_(tmp_id1, tmp_id2)])
else: # method == "u"
iter_vals[i] = mmd_unbiased(K[np.ix_(tmp_id1, tmp_id1)],
K[np.ix_(tmp_id2, tmp_id2)],
K[np.ix_(tmp_id1, tmp_id2)])
p_value = (np.sum(iter_vals >= stat) + 1) / (mc_iter + 1)
return {
'statistic': {'MMD': stat},
'p_value': p_value,
'alternative': "two distributions are not equal",
'method': "Kernel Two-sample Test with Maximum Mean Discrepancy"
}
We can create an example to explain how to use the code:
np.random.seed(42)
# Create Beta distributions and generate samples
x = stats.beta(2, 5).rvs(15)
y = stats.beta(5, 5).rvs(15)
# Points for plotting the density curves
z = np.linspace(-0.5, 1.5, 100)
density_x = stats.beta(2, 5).pdf(z)
density_y = stats.beta(5, 5).pdf(z)
# Plot
plt.scatter(x, np.zeros_like(x), marker='+')
plt.plot(z, density_x)
plt.plot(z, density_y)
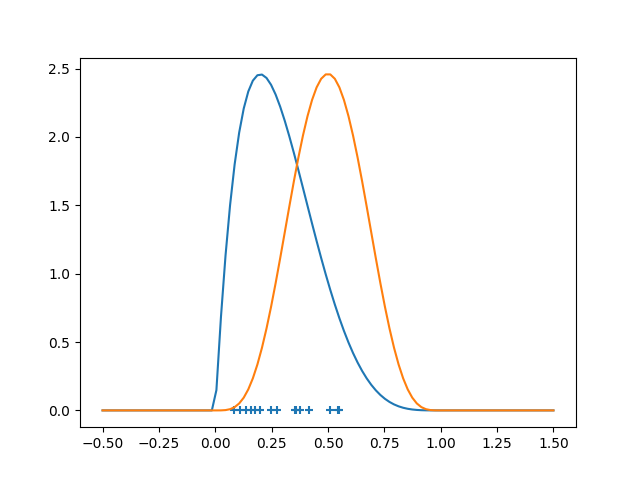
Now we analyse the data:
# Reshape the data to be 2D arrays
x = x.reshape(-1, 1) # Shape becomes (15, 1)
y = y.reshape(-1, 1) # Shape becomes (15, 1)
# Combine data and compute distance matrix
combined_data = np.vstack([x, y])
distances = squareform(pdist(combined_data))
# Compute median distance for kernel bandwidth
sigma = np.median(distances)
print(f"Using median bandwidth: {sigma:.4f}")
# Build Gaussian kernel matrix with scaled distances and small regularization
kernel_matrix = np.exp(-distances**2 / (2 * sigma**2))
kernel_matrix += 1e-15 * np.eye(len(kernel_matrix)) # Small regularization
# Create labels
labels = np.array([1]*len(x) + [2]*len(y))
# Run the test
result = mmd2test(kernel_matrix, labels)
print("\nTest Results:")
print(f"MMD statistic: {result['statistic']['MMD']:.6f}")
print(f"p-value: {result['p_value']:.6f}")
Which gives
Test Results:
MMD statistic: 0.416771
p-value: 0.003000